Research Summary
Our laboratory is integrating cutting-age basic science together with clinical data in order to identify novel genes and cellular processes involved in tumor cell progression and response to target therapies.

Growth factor signalling
Research Summary
Our laboratory is integrating cutting-age basic science together with clinical data in order to identify novel genes and cellular processes involved in tumor cell progression and response to target therapies.

Growth factors acting through receptor tyrosine kinases (RTKs) of ERBB family, integrate a network of multiple inputs, such as ligands of cell-surface receptors; and produce multiple outputs, from gene expression and cellular activities, including motility, proliferation and death. With the “omics era”, the complexity of both intra e inter cellular interactions become manifest, thus our mission is trying to understand the special laws of nature modelling biological systems.
Transcription networks are designed with specific timescales. In response to a stimuli signal-transduction networks operate immediately, by transmitting the input to several Transcription Factors and once on the DNA, they are engaged in the production of target genes.
Steroid hormones activating nuclear localized receptors, are able to bypass the transduction networks, thus the timing of their activities result faster and relatively simple. Nevertheless, in a living organism, these two systems interact massively, with major consequences for body homeostasis and cell activities.
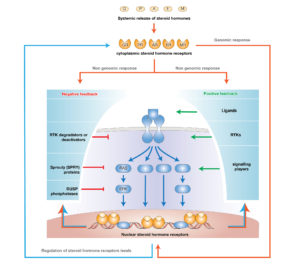
This line of research focuses on the transcriptional modifications resulting from steroid hormones and growth factors interaction. 
We previously showed that capturing epigenetic mechanisms and transcriptional control of several feedback regulatory loops, steroid hormones (GCs) gained the ability to robustly inhibit EGF induced signals leading to cell migration. Specifically, GCs enhance auto-inhibitors of EGF signaling and inhibit expression of auto-stimulators of the same pathway.
We are studying the mechanisms driving this interaction in order to optimize the response to EGFR target therapy, in several biological backgrounds, either mammary epithelial tissues and gastrointestinal tissues.
EGF Cell migration under EGF stimulation
16_EDX Delayed cell migration under EGF plus Dex stimulation
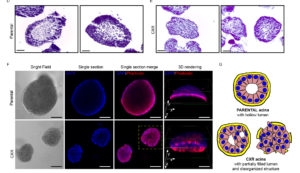
Resistance to EGFR blockade may have a genetic basis as well as activation of downstream or parallel signaling pathways that substitute for EGFR inhibition. This process relies on plastic, reversible traits induced by drug pressure, such as compensatory activation of biochemical feedback circuits and transcriptional modifications.
In this project, we aim to explore the plastic phenotype of cellular adaptation to prolonged drug treatment.
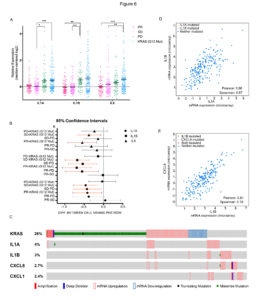
We found that resistance to EGFR targeting drugs results in the up-regulation of a signature of inflammatory cytokines, namely IL1A, IL1B and IL8. The association between reduced sensitivity to anti-EGFR antibodies and increased expression of this inflammatory signature was confirmed in patients’ data and specimens.
We employ monolayer and 3D-spheroid growing cells to decipher the mechanisms by which an inflammatory microenvironment might impair cetuximab efficacy in patients.
Roth L, Lindzen, Sas-Chen A, Noronha A, Sheffer M, Mancini M, Enuka Y, Tarcic G, Pines G, Lauriola M, Nevo N, Porat Z, Sevilla-Sharon M, Carvalho S, Ziv T, Domany E, Admon E, Caldas C and Yarden Y. (2018).SILAC identifies LAD1 as a filamin-binding regulator of actin dynamics in response to EGF and a marker of aggressive breast tumors. Science Signaling 2018.
Rodia MT, Solmi R, Pasini F, Nardi E, Mattei G, Ugolini GP, Ricciardiello L, Strippoli PL, Miglio R, Lauriola M (2018). LGALS4, CEACAM6, TSPAN8, and COL1A2: Blood Markers for Colorectal Cancer-Validation in a Cohort of Subjects With Positive Fecal Immunochemical Test Result. Clin Colorectal Cancer. 2017 Dec 12
Enuka Y, Feldman ME, Chowdhury A, Srivastava S, Lindzen M, Sas-Chen A, Massart R, Cheishvili D, Suderman MJ, Zaltsman Y, Mazza CA, Shukla K, Körner C, Furth N, Lauriola M, Oren M, Wiemann S, Szyf M, Yarden Y. (2017). Epigenetic mechanisms underlie the crosstalk between growth factors and a steroid hormone. Nucleic Acids Res. 2017 Dec 15;45(22):12681-12699. doi: 10.1093/nar/gkx865.
Gelfo, V., Rodia, M. T., Pucci, M., Dall’Ora, M., Santi, S., Solmi, R., Lauriola, M. (2016).
A module of inflammatory cytokines defines resistance of colorectal cancer to EGFR inhibitors. Oncotarget. https://doi.org/10.18632/oncotarget.12354
Li, Y., Lauriola, M., Kim, D., Francesconi, M., D’Uva, G., Shibata, D., … Cheng, J. Q. (2016).Adenomatous polyposis coli (APC) regulates miR17-92 cluster through β-catenin pathway in colorectal cancer. Oncogene, 35(35), 4558–4568. https://doi.org/10.1038/onc.2015.522
D’Uva, G., & Lauriola, M. (2016).
Shaping ERBB Signalling by Steroid Hormones. J Steroids Hormon Sci, 7(1000.166), 2.
Rodia, M. T., Ugolini, G., Mattei, G., Montroni, I., Zattoni, D., Ghignone, F., … Solmi, R. (2016).Systematic large-scale meta-analysis identifies a panel of two mRNAs as blood biomarkers for colorectal cancer detection. Oncotarget. https://doi.org/10.18632/oncotarget.8108
Carvalho, S., Lindzen, M., Lauriola, M., Shirazi, N., Sinha, S., Abdul-Hai, a, … Yarden, Y. (2015).An antibody to amphiregulin, an abundant growth factor in patients’ fluids, inhibits ovarian tumors. Oncogene, 35(October 2014), 1–10. https://doi.org/10.1038/onc.2015.93
Enuka, Y., Lauriola, M., Feldman, M. E., Sas-Chen, A., Ulitsky, I., & Yarden, Y. (2015).Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Research, gkv1367-. https://doi.org/10.1093/nar/gkv1367
Kedmi, M., Ben-Chetrit, N., Körner, C., Mancini, M., Ben-Moshe, N. B., Lauriola, M.,Yarden, Y. (2015).EGF induces microRNAs that target suppressors of cell migration: miR-15b targets MTSS1 in breast cancer. Science Signaling, 8(368), ra29-ra29.
D’Uva, G., Aharonov, A., Lauriola, M., Kain, D., Yahalom-Ronen, Y., Carvalho, S., … Tzahor, E. (2015).ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nature Cell Biology, 17(5), 627–638.
Cohen-Dvashi, H., Ben-Chetrit, N., Russell, R., Carvalho, S., Lauriola, M., Nisani, S., … Yarden, Y. (2015).Navigator-3, a modulator of cell migration, may act as a suppressor of breast cancer progression. EMBO Molecular Medicine, 7(3), 299–314. https://doi.org/10.15252/emmm.201404134
Ben-Chetrit, N., Chetrit, D., Russell, R., Korner, C., Mancini, M., Abdul-Hai, A., … Yarden, Y. (2015).Synaptojanin 2 is a druggable mediator of metastasis and the gene is overexpressed and amplified in breast cancer. Science Signaling, 8(360), ra7. https://doi.org/10.1126/scisignal.2005537
D’Uva, G., & Lauriola, M. (2015).
Towards the emerging crosstalk: ERBB family and steroid hormones. Seminars in Cell and Developmental Biology. Academic Press.
Lauriola, M., Enuka, Y., Zeisel, A., D’Uva, G., Roth, L., Sharon-Sevilla, M., … Yarden, Y. (2014).Diurnal suppression of EGFR signalling by glucocorticoids and implications for tumour progression and treatment. Nature Communications, 5, 5073.
Peña-Altamira, E., Polazzi, E., Moretto, E., Lauriola, M., & Monti, B. (2014).
The transcription factor CCAAT enhancer-binding protein β protects rat cerebellar granule neurons from apoptosis through its transcription-activating isoforms. European Journal of Neuroscience, 39(2), 176–185.
D’Uva, G., Bertoni, S., Lauriola, M., De Carolis, S., Pacilli, A., D’Anello, L.,Storci, G. (2013).Beta-catenin/HuR post-transcriptional machinery governs cancer stem cell features in response to hypoxia. PLoS ONE, 8(11).
Zeisel, A., Yitzhaky, A., Koerner, C., Lauriola, M., Cohen-Dvashi, H., Köstler, W. J., … Domany, E. (2013).qCMA: a desktop application for quantitative collective cell migration analysis. Journal of Biomolecular Screening, 18(3), 356–60.
Pradeep, C.-R., Köstler, W. J., Lauriola, M., Granit, R. Z., Zhang, F., Jacob-Hirsch, J., … Yarden, Y. (2012).
Modeling ductal carcinoma in situ: a HER2-Notch3 collaboration enables luminal filling. Oncogene, 31(7), 907–17. https://doi.org/10.1038/onc.2011.279
Pradeep, C.-R., Zeisel, A., Köstler, W. J., Lauriola, M., Jacob-Hirsch, J., Haibe-Kains, B., … Yarden, Y. (2012).
Modeling invasive breast cancer: growth factors propel progression of HER2-positive premalignant lesions. Oncogene, 31(31), 3569–83. https://doi.org/10.1038/onc.2011.547
Rivetti, S, Lauriola, M., Voltattorni, M, Bianchini, M, Martini, D, Ceccarelli, C, Palmieri, A, Mattei, G., Franchi, M., Ugolini, G, Rosati, G., Montroni, I, Taffurelli, M., Solmi, R. (2011).
Gene expression profile of human colon cancer cells treated with cross-reacting material 197, a diphtheria toxin non-toxic mutant. International Journal of Immunopathology and Pharmacology, 24(3), 639–649.
Lauriola, M., Ugolini, G., Rivetti, S., Nan??, S., Rosati, G., Zanotti, S., … Solmi, R. (2011).IL23R, NOD2/CARD15, ATG16L1 and PHOX2B polymorphisms in a group of patients with Crohn’s disease and correlation with sub-phenotypes..International Journal of Molecular Medicine, 27(3), 469–477. https://doi.org/10.3892/ijmm.2010.591
Zwang, Y., Sas-Chen, A., Drier, Y., Shay, T., Avraham, R., Lauriola, M., … Yarden, Y. (2011).Two phases of mitogenic signaling unveil roles for p53 and EGR1 in elimination of inconsistent growth signals. Molecular Cell, 42(4), 524–535.
Lauriola, M., Ugolini, G., Rosati, G., Zanotti, S., Montroni, I., Manaresi, A., … Solmi, R. (2010).Identification by a Digital Gene Expression Displayer (DGED) and test by RT-PCR analysis of new mRNA candidate markers for colorectal cancer in peripheral blood. International Journal of Oncology, 37(2), 519–525.
Solmi, R., Lauriola, M., Francesconi, M., Martini, D., Voltattorni, M., Ceccarelli, C., … Strippoli, P. (2008).Displayed correlation between gene expression profiles and submicroscopic alterations in response to cetuximab, gefitinib and EGF in human colon cancer cell lines. BMC Cancer, 8, 227.
 Dr. Mattia Lauriola, Ph.D
Dr. Mattia Lauriola, Ph.D
Principal Investigator
mattia.lauriola2@unibo.it
+39 0512094118
 Gabriella Mattei
Gabriella Mattei
Laboratory Technician
gabriella.mattei@unibo.it
+39 0512094126

Enea Ferlizza
Laboratory Technician
enea.ferlizza2@unibo.it
+39 0512094115

Donatella Romaniello, PhD
Junior Assistant Researcher
donatella.romaniello@unibo.it
+39 0512094087
 Valerio Gelfo, Msc
Valerio Gelfo, Msc
Postdoctoral Fellow
valerio.gelfo2@unibo.it
+39 0512094102
 Martina Mazzeschi
Martina Mazzeschi
Researcher
martina.mazzeschi2@unibo.it
+39 0512094102
 Michela Sgarzi
Michela Sgarzi
Researcher
Michela.Sgarzi2@unibo.it
+39 0512094102

Alessandra Morselli
PhD student
alessandra.morselli4@unibo.it
+39 0512094102
 Cinzia Girone
Cinzia Girone
PhD student
cinzia.girone2@unibo.it
+39 0512094102
 Federica Pagano
Federica Pagano
PhD student
federica.pagano2@unibo.it
+39 0512094102
 Elisa Montacci
Elisa Montacci
PhD student
elisa.montacci2@unibo.it
+39 0512094102
 Francesco Borrelli
Francesco Borrelli
PhD student
francesco.borrelli3@unibo.it
+39 0512094102
Varun Shankar
PhD student
varun.shankar3@unibo.it
+39 0512094102
Chiara Miroglio
Anna Francia
Jasmijn Timmermans

Department of Experimental, Diagnostic and Specialty Medicine – DIMES
University of Bologna
Bologna, Italy
mattia.lauriola2@unibo.it